As I mentioned the other week, I’m analyzing participation rates of gender, ethnic, and racial minorities in cancer clinical trials concluded between 2002-2012. I’m working with 3 datasets from which I’ve been gleaning the information relevant for my analysis. I started out by looking at the Aggregate Analysis of ClinicalTrials.gov (AACT), made available by CTTI in pipe-delimited and Oracle .dmp formats. Eric graciously re-assembled the .dmp data into a usable Oracle database for me. I then used a program called DBVisualizer to do some basic Structured Query Language (SQL) queries.
I’ve never worked with relational databases before, so I started by opening the data dictionary Excel file that’s packaged with AACT. This file contains not just the dictionary of variables, but also a graphical representation of the database schema that looks a bit like a flowchart. A schema is a map of a dataset’s structure that explains the relationships between tables. Once I realized which tables and variables were actually relevant for my purposes (thankfully, not all of them!), I set about learning some rudimentary SQL to help me retrieve the specific information I’m after. The tutorials at Vertabelo were pretty simple to follow and I highly recommend them.
At first, I was a bit puzzled at how little usable information I was getting from my initial queries. I couldn’t figure out why condition_browse.mesh_term like '%cancer%' wasn’t grabbing the entire list of cancer clinical trials for me. Then I learned that AACT data is indexed using Medical Subject Headings (MeSH), the U.S. National Library of Medicine’s thesaurus for indexing of medical articles. MeSH codes each cancer as a type of neoplasm. So breast cancer is going to appear in a MeSH-indexed database as “breast neoplasm.” What a difference one string makes! Since there are a dizzying number of cancer types, I chose to focus on 5 common ones: breast, prostate, lung, ovarian, and colorectal. I also didn’t account for the date information to be coded as strings rather than…dates. So my queries have been a little clunky, but effective.
I’m also working with the National Cancer Institute’s Surveillance, Epidemiology, and End Results (SEER) Program cancer registry data, which provides cancer case information and incidence rates for a sample of the U.S. population (roughly equivalent to 28% of the country). I had to fill out a request and get permission to access it, which felt moderately fancy. NCI helpfully bundles the data with a special statistical package called SEERStat designed to facilitate epidemiological research. The software is an interface by which users can tailor exactly what types of tables (called matrices) they’re after. It’s very customizable, and I created some user-defined variables to reflect the time period and cancer types of interest, then executed a session and got a really neat table organized perfectly for my needs. I like that the SEERStat variables I’ve created are shareable, and I’ll be including them with my research findings in the interests of transparency. Here’s a snippet of an executed SEER*Stat session:
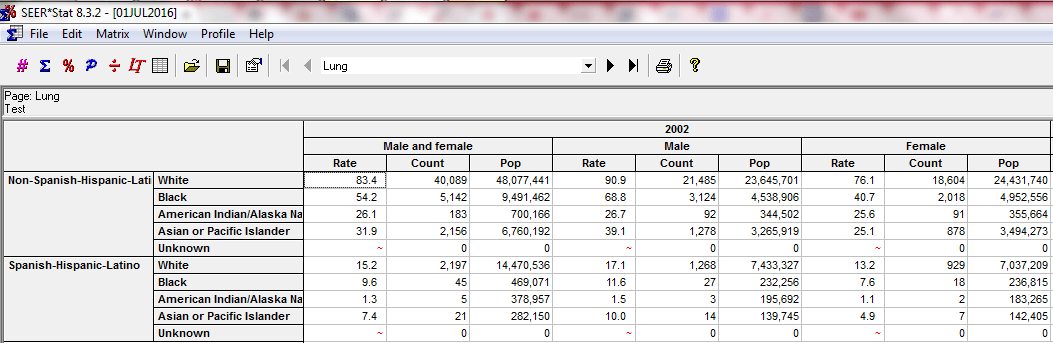
The third and final source of data for my project originates from the U.S. Census bridged-race population estimates. At first, I went and downloaded each .txt file for each year separately. Then I discovered an online tool called CDC WONDER that provides a breezy online interface for querying the most recent population estimates by factors like age, race, ethnicity, gender, etc. I was easily able to get all the population data I needed, within seconds.
By applying the cancer incidence rates obtained via SEER*Stat to the Census numbers, I’ve been able to estimate the national incidence numbers for each cancer, for each gender/ethnic/racial group. Eric and I are working to clean up the last bit of AACT data and then I can start doing some more advanced calculations with my new friend R!